Small World Work
HCC honors biology class joins global search for new antibiotics

The process begins with dirt. A small sample will do. It doesn't really matter where it's from.
In the lab, the soil — that's a more scientific word — is diluted and streaked over the surface of an agar-filled petri dish and incubated.
Next, the students wait to see what types of microbes grow.
"Our hope is that they haven't been found before, because discovering a new bacteria in this classroom would be amazing," said Justin Roberts of South Hadley, a student enrolled during the fall semester in Prof. Carolyn Wetzel's "Introduction to Cell Functions" course at Holyoke Community College.
Typically, Bio 107, also known as General Biology I, runs as a standard, introductory laboratory science. Wetzel's class is different. It's an honors biology course connected to a global research project called the Small World Initiative.
The Small World Initiative enlists college and high school science students all over the world to literally search their local dirt for previously undiscovered strains of antibiotic-producing bacteria. The premise: The more people who look the more likely they will be to find.
"It's a crowd-sourced research project," said Wetzel. "Pharmaceutical companies aren't really looking for new antibiotics, and there's a lot of resistance building up to the antibiotics we already have. It's predicted that by the year 2050 they'll be more people dying from resistant strains of bacterial infections than from heart disease and some of the other big killers — if we don't start finding new ones."
The Small World Initiative started at Yale University in 2012 and now includes some 250 partner schools in 38 states and 14 countries. Wetzel attended a five-day training session last July at the University of Connecticut, where she learned SWI laboratory techniques and teaching methods.
"There are Small World partners in Spain and South America," she said. "It's in a lot of places, but not that many in Massachusetts yet. We're the only community college in Massachusetts doing this."
Wetzel's class covers the same basic lecture material as the other Bio 107 sections, focusing on a new topic each week, such as osmosis or enzymes. The lab portion, though, is "completely different," she said.
In regular Bio 107, students conduct prepared laboratory exercises based on that week's lecture material. The results of experiments are often predetermined. No matter what the outcome, next week you move on.
"This is a research class," Wetzel said during one Friday lab. "We have a research goal and we work on it through the semester. If something doesn't work one week, you have to figure it out and make it work because you can't go forward without that step."
That approach is what attracted many of her students to the honors class.
"I think this gives us more of an opportunity to be involved directly in science, rather than just sitting in a lecture and listening to it," said Ariana Shannon of Becket. "I think it's interesting how no one is looking for new antibiotics to fight illnesses. It's cool that college students get to look for them instead."
The first step in the process of finding new antibiotics is to identify bacteria that produce them. And that begins with dirt.
"That's where a lot of our antibiotics come from," Wetzel said.
At the beginning of the semester, she sent her 16 students out to scoop soil. Some brought in samples from home. Some dug around campus.
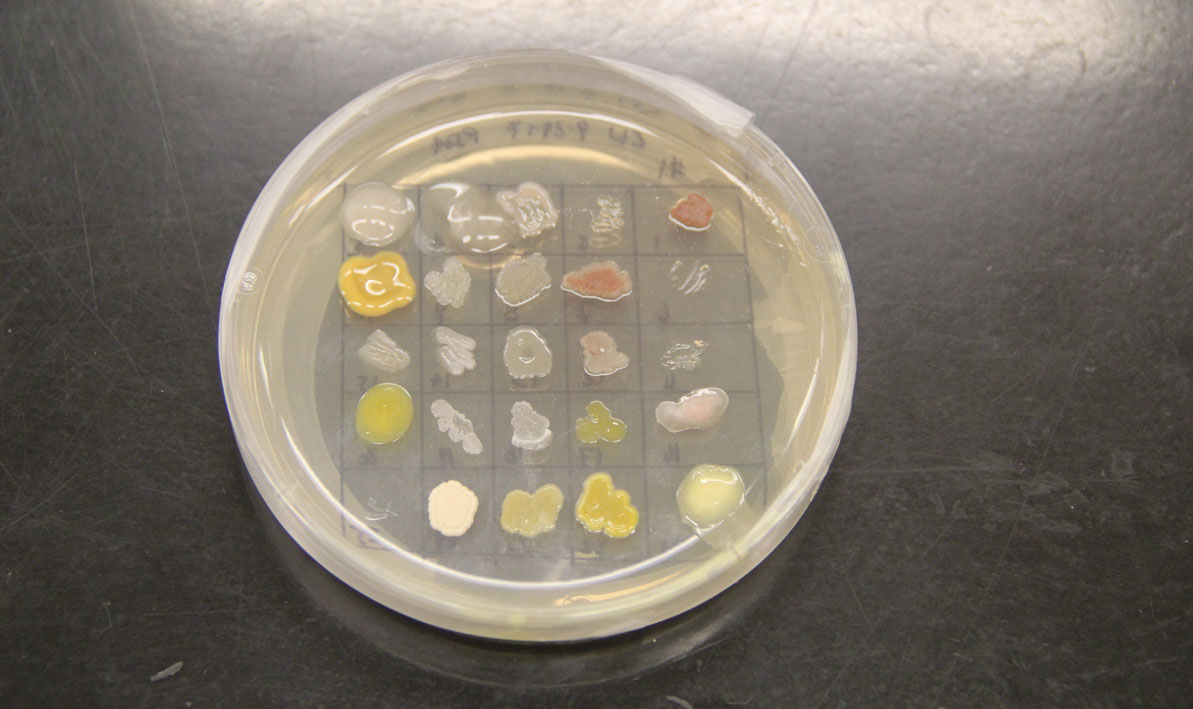
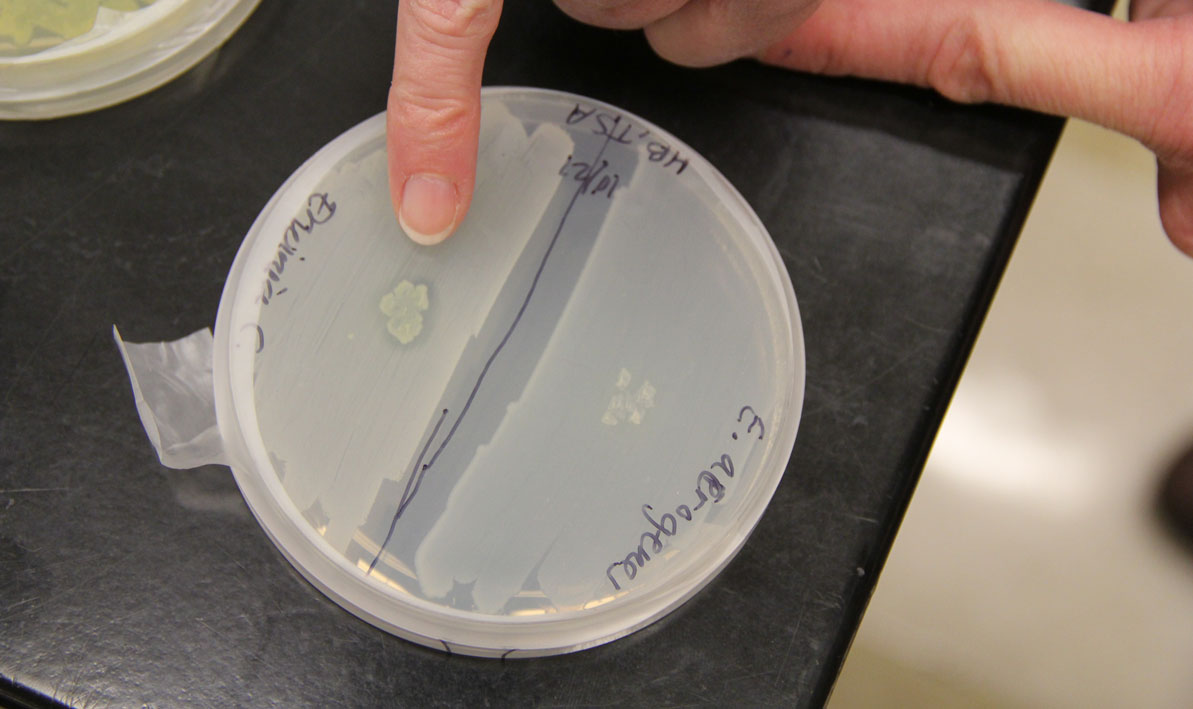
Students isolated bacteria from those samples and then tested them against safe strains of their more pathogenic cousins with names like Staphlococcus epidermis, Escherichia coli and Enterococcus raffinosus, careful to note the ones where a clear halo surrounded the cultures like a protective moat.
"That tells us the bacteria they isolated is producing an antibiotic — a chemical that can do the killing," she said.
After that, the students extracted strands of DNA from the antibiotic-producing bacteria and then used a process called a polymerase chain reaction to replicate that DNA in a test tube. The DNA samples were sent to Yale for genetic sequencing, with the results compared against an international database of known bacteria.
"It's the same technique that's used for determining DNA evidence at crime scenes, only we're doing it with bacteria," Wetzel said.
While many of the students did find antibiotic-producing bacteria during the semester, Wetzel said, results have not yet determined whether they have been previously undiscovered. At least two of her students will continue their research next semester as independent study projects.
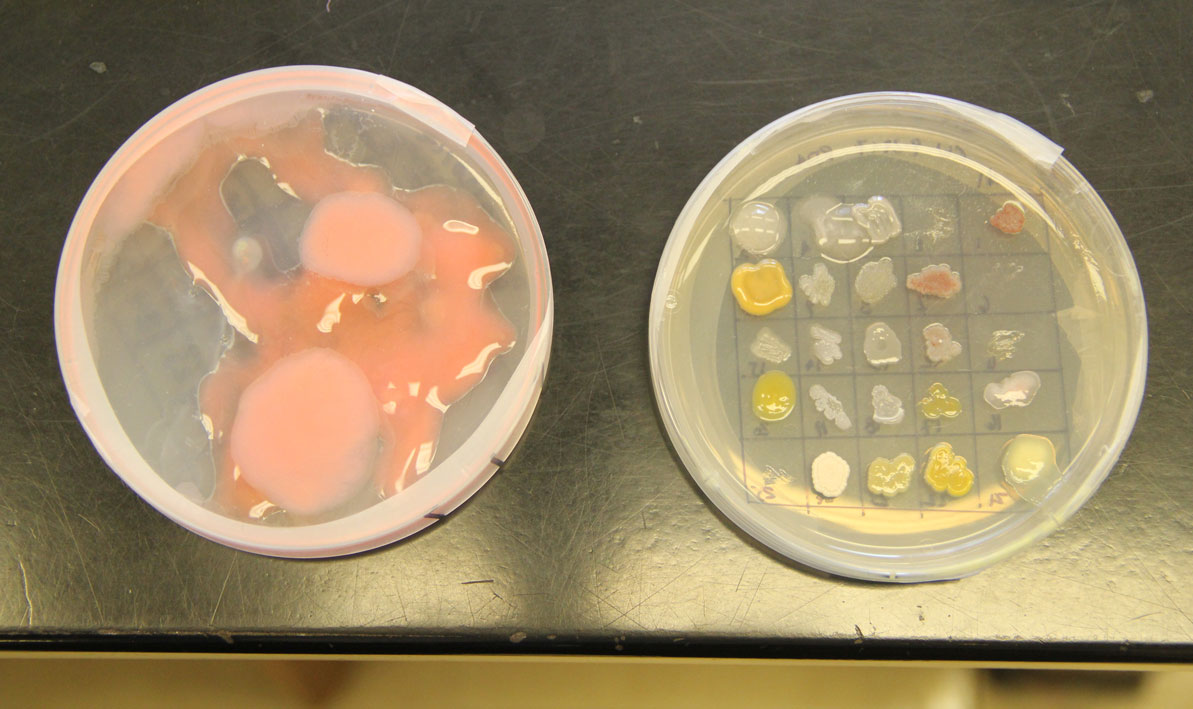
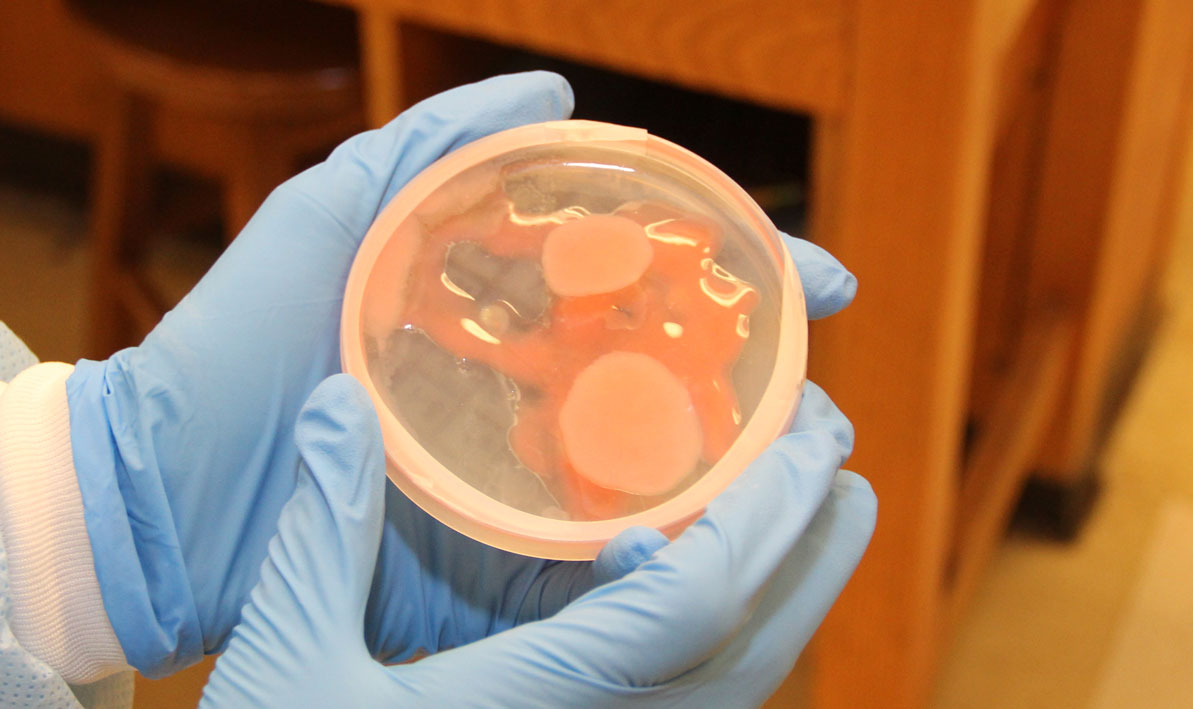
For all the excitement about the potential for finding new disease-fighting antibiotics, a development that caused a significant buzz was an microorganism that completely swamped one petri dish like a blob of pink bubble gum, smothering all the other cultures in its path.
"We don't know what that is," Wetzel said.
STORY and PHOTOS by CHRIS YURKO: (Thumbnail) HCC honors biology student Justin Roberts of South Hadley uses a micro-pipette to deposit strands of DNA into test tube. (Above) HCC honors biology lab partners Shayla Fiedler from Longmeadow and Ariana Shannon from Beckett look for antibiotic activity in a petri dish filled with different strains of bacteria.